Portfolio for Data Science

30+ projects completed in R and Python demonstrate my expertise in data cleaning, analysis, visualization, and machine learning, serving as a testament to my growth and competency in the dynamic field of data science. Learn more
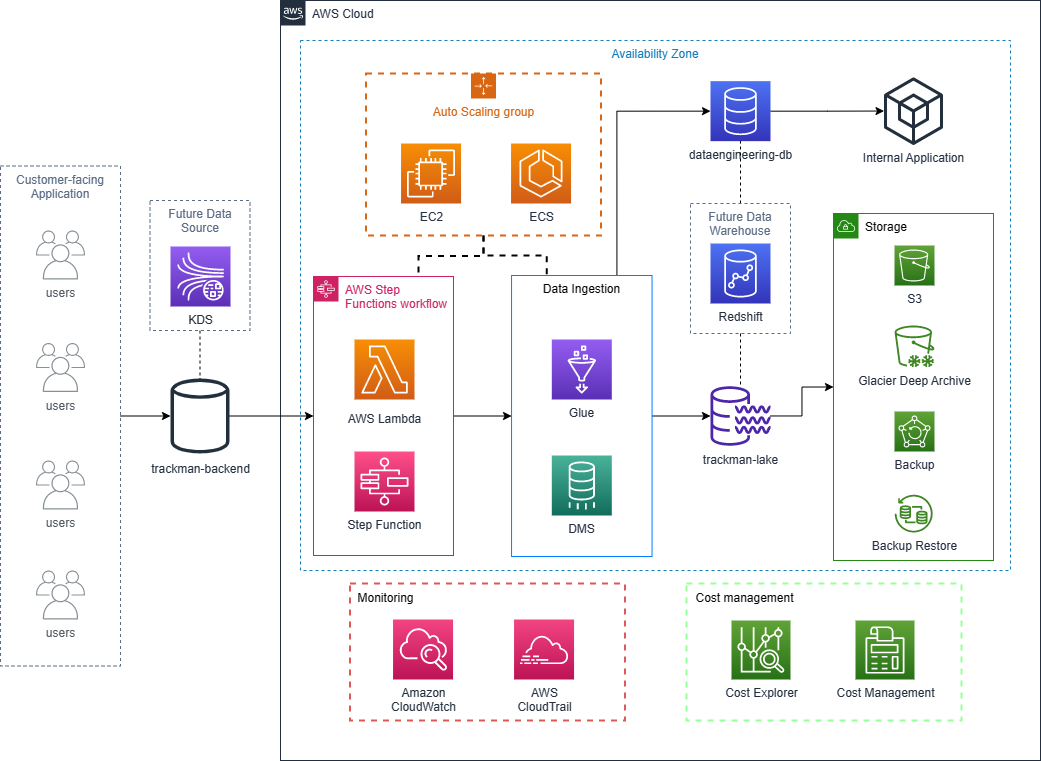
Data Infrastructure Design in AWS
The TrackMan Data Engineering System Design Challenge aims to gauge the familiarity with designing cloud based applications and solutions. This challenge is as much about the ability to communicate and justify the decisions as it is the specifics of the design itself.
Click the diagram to find the design details.
GSoC 2023
Google Summer of Code is a global, online program focused on bringing new contributors into open source software development. GSoC Contributors work with an open source organization on a 12+ week programming project under the guidance of mentors.
Here are the projects I am currently contributing:
FURY
Free Unified Rendering in Python.
A software library for scientific visualization in Python.
Open Source https://github.com/fury-gl/fury
Scientific axes for FURY
The ability to effectively display scientific data requires a correspondence between the actual and displayed scales and the use of axes to indicate such scales. Many visualization frameworks, such as d3 and Matplotlib, include a variety of ways to generate axes and frames based on the domain of the plotted data. In 3D, axes can be represented by lines or grid planes, and more work is needed to make them practical (such as displaying shadows or lines perpendicular to the grid planes).
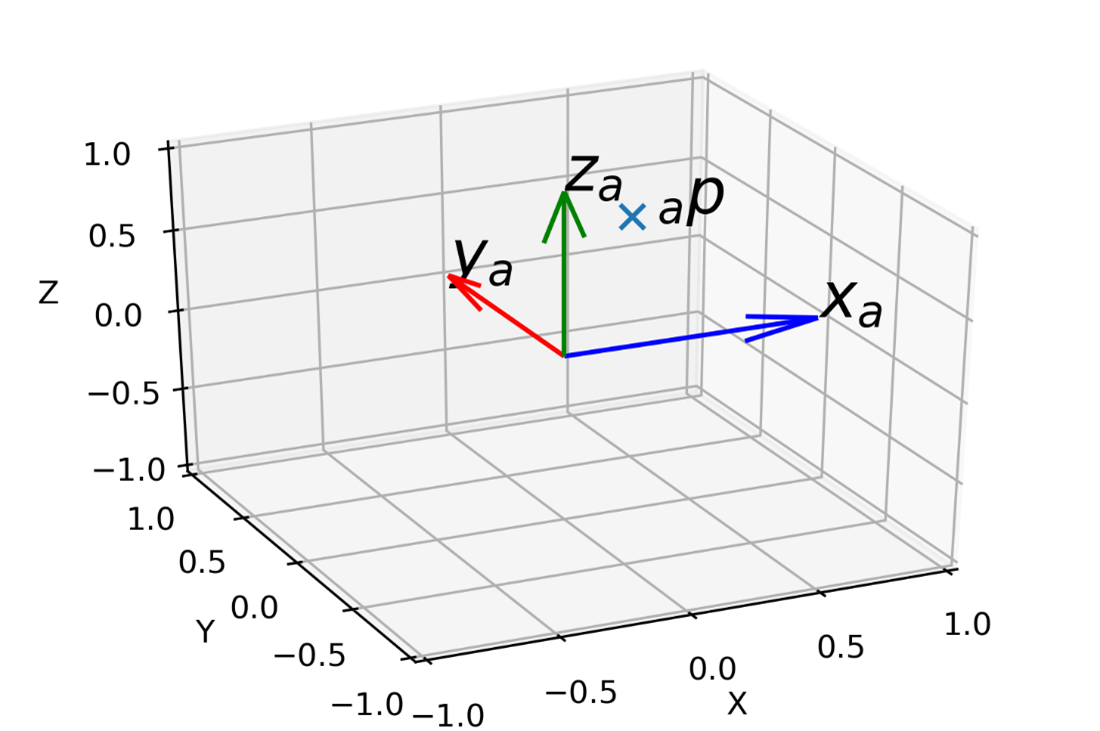
This project aims to implement a comprehensive set of actors to display dynamic scientific axes, both in 2D and 3D. This includes the development of a user-friendly API for defining ranges, linear and non-linear transformations, and customizing axes with colors, widths, labels, ticks, and more. Additionally, the project will implement high-quality 3D grids with support for shadows and orthogonal lines, as well as 2D axes, with improved heuristics for distributing labels and ticks. The axes will be animatedly through the keyframe animation API, allowing for interpolation between changes. The implementation of these features will significantly enhance the ability of FURY to produce high-quality scientific visualizations that accurately communicate data to the intended audience.
d-SEAMS

Deferred Structural Elucidation Analysis for Molecular Simulations.
An organization centered around growing the molecular dynamics post processing toolkit called d-SEAMS
Open Source https://github.com/d-SEAMS/seams-core
Overview
d-SEAMS is an engine meant to interface with molecular dynamics trajectories. Like those generated by LAMMPS, the primary software of choice for most d-SEAMS users. As a post-processing tool, for gaining insights into the dynamics systems simulated, it is often best to reduce the gap between trajectories and data. To this end, the current design of d-SEAMS is controlled by a YAML file per analysis, along with a scripting engine, which is currently in Lua. This scripting engine is flexible, and prevents erroneous mixing of unsupported code for the most part, however, Lua is not necessarily well known to most practicing scientists. To this end, the scripting engine will be replaced with a Python embedded interpreter. Rather than writing Python-C code directly to manage the interface, we plan to use the the Pybind11 project.
Connecting to the Python Molecular Dynamics community
Workflows have taken over the high-performance computing landscape, for example, those executed within Jupyter (e.g. PyIron) or without (e.g. AiiDA). These tools often have sophisticated or baroque object relation management models (e.g. PostGreSQL in AiiDA). However, they all also provide an interface through Python.
The goal of this project is to integrate through Pybind11 to these external codes namely:
cclibfor transforming data and inputs from different code-basesasefor integrating with user-designed rapid prototyping tools, e.g. we would like to be able to integrate seamlessly with the existingasetoolset, to (e.g.) run an NEB (nudged-elastic-band) and also track the shapes during the processi-piwhich can run additional dynamical simulations (in Python)
A stretch goal here is to convert the internal data structures to an SQL data-base for better integration with AiiDA, however this will need more design inputs.